Technologies of CLST
Incorporation of synthetic amino acids into proteins at specific sites
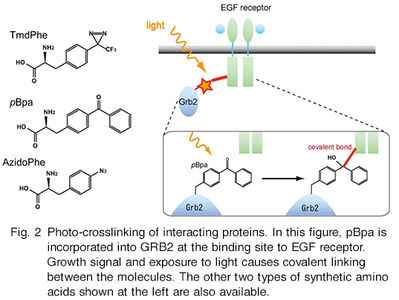
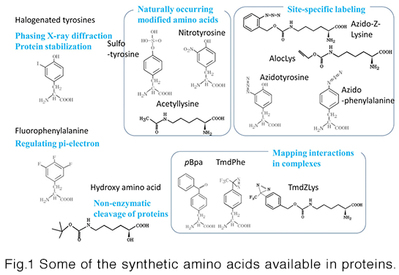 This technology is about site-specific incorporation of synthetic amino acids into proteins in animal cells andE. coli cells. It can be applied to (i) synthesize proteins having naturally-occurring modifications (acetyllysine, sulfotyrosine, and nitrotyrosine) at specific sites, (ii) site-selectively modify proteins through incorporation of bio-orthogonally reactive groups, such as azido and alkyne, (iii) site-selectively incorporate photo-reactive amino acids, such as pBpa, into proteins for forming covalent bonds between interacting proteins, and (iv) enhance structural stability of proteins through halogenations of selected tyrosine positions. We can provide the following host systems to incorporate synthetic amino acids into proteins.
This technology is about site-specific incorporation of synthetic amino acids into proteins in animal cells andE. coli cells. It can be applied to (i) synthesize proteins having naturally-occurring modifications (acetyllysine, sulfotyrosine, and nitrotyrosine) at specific sites, (ii) site-selectively modify proteins through incorporation of bio-orthogonally reactive groups, such as azido and alkyne, (iii) site-selectively incorporate photo-reactive amino acids, such as pBpa, into proteins for forming covalent bonds between interacting proteins, and (iv) enhance structural stability of proteins through halogenations of selected tyrosine positions. We can provide the following host systems to incorporate synthetic amino acids into proteins.
Animal cells
Synthetic amino acids can be site-specifically incorporated into proteins in mammalian and insect S2 cells. We usually use CHO and HEK cells, and have little information concerning the availability of other cell types. The mechanism of the incorporation of such amino acids is as follows. UAG codon is allotted to a synthetic amino acid, while at the same time the codon still works as a stop codon. This ambiguous codon assignment results in the production of two types of molecules: the full-length protein in which the synthetic amino acid is incorporated in response to UAG codon within the gene, and the truncated protein whose translation was terminated at the in-frame UAG codon. In most cases, the yield of full-length proteins with synthetic amino acids is 5 to 20% of that of the wild-type full-length proteins with no such amino acids. The efficiency of incorporation is not the same among positions in the proteins. Some sites do not allow their incorporation. It is impractical to incorporate synthetic amino acids at more than one site. To incorporate these amino acids, you need to transform cells with plasmids carrying genes necessary for the purpose, and then supplement the growth media with the amino acids. Request the plasmids. You will get them on the MTA basis. Purchase the cells and the amino acid on your own.
Normal E .coli cells
The mechanism of incorporation is similar to the aforementioned animal cells. We will get full-length proteins with synthetic amino acids and truncated products. The ration between these molecules is usually much better than in the animal cells, and more than 20% of the products are expected to be contain synthetic amino acids. There is a difference than the animal cell system. The incorporation efficiency is strongly influenced by the codon context effect. It is generally much easier to incorporate the amino acids in response to UAG just preceding a codon starting with a purine than UAG just preceding a codon starting with a pyrimidine. This means that you can more efficiently incorporate the amino acids just before glycine (GGN), arginine (AGR) etc than phenylalaine (UUY), proline (CCN) etc. As in the animal cell system, you need to transform E. coli with the plasmids and supplement the media with the amino acids. We can share the plasmids on the MTA basis, but cannot afford to provide a synthetic amino acid.
Engineered E. coli or RFzero
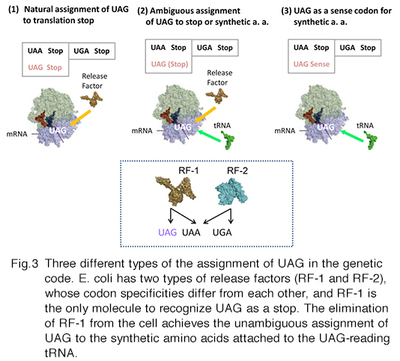
We have developed specialized E. coli cells, called RFzero, in which the UAG codon has been reassigned to synthetic amino acids and lost its original assignment to translation termination. Just as the other sense codon, UAG can appear multiple times within a gene without terminating protein synthesis. Thus, the efficiency of incorporation is almost 100%, enabling the incorporation at multiple specific sites. RFzero strains require the supplementation of synthetic amino acids in the growth media. For example, the RFzero-azf strain, in which UAG is redefined as a sense codon specifying 4-azidophenylalanine, requires this amino acid to grow. You only have to transform the strain with a recombinant gene with (a) in-frame UAG codons, to synthesize the recombinant protein with azidophenylalanine(s). We will share RFzero strains on the basis of MTA and the agreement on the transfer of GMO. We have improved the strains so that a wide variety of synthetic amino acids are now available for the UAG reassignment. The results will be published soon.
This paragraph is for the reader interested in the mechanism for the UAG reassignment. In bacteria, the UAG stop codon is recognized by release factor 1 (RF-1). Since there are about 300 open reading frames ending with UAG codons in the E. coli genome, RF-1 is an indispensable cellular factor. We have found that RF-1 can be eliminated from the bacteria, if 8 important genes are engineered to end with UAA in place of UAG, and UAG-reading tRNA is expressed in the cell. After the elimination of the factor, the meaning of UAG unambiguously corresponds to the amino acid identity of the UAG-reading tRNA. A specific pair of UAG-reading tRNA and an aminoacyl-tRNA synthetase variant has been developed by researchers for each of >100 synthetic amino acids. In principle, these amino acids are available in RFzero. The improved RFzero is for enhancing this availability. Another advantage of RFzero is that it can be created rapidly, and thus several E. coli strains have been employed as parents of RFzero. Among them, there is BL21(DE3), most frequently used as a host cell for synthesizing recombinant proteins in a large scale. BL21(DE3)-based RFzero has proved useful to efficiently incorporate azidophenylalanine, halogenated tyrosines, and nitrotyrosines.
References
[Photo-crosslinking]
-
Hino, N., Okazaki, Y., Kobayashi, T., Hayashi, A., Sakamoto, K., Yokoyama, S., “Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid”, Nat. Methods 2, 201-206 (2005)
-
Hino, N., Oyama, M., Sato, A., Mukai, T., Iraha, F., Hayashi, A., Kozuka-Hata, H., Yamamoto, T., Yokoyama, S. & Sakamoto, K. Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. J. Mol. Biol. 406, 343-53 (2011)
[Probing cation-pi interactions involved in the catalytic activity of an enzyme]
-
Morikubo, N., Fukuda, Y., Ohtake, K., Shinya, N., Kiga, D., Sakamoto, K., Asanuma, M., Hirota, H., Yokoyama, S., Hoshino, T., “Cation-π interaction in triterpene biosynthesis revealed by incorporating unnatural amino acids into the catalytic site of a squalene cyclase”, J. Am. Chem. Soc. 128, 13184-13194 (2006)
[Site-selective modification of proteins]
-
Yanagisawa, T., Ishii, R., Fukunaga, R., Kobayashi, T., Sakamoto, K., Yokoyama, S., “Multi-step engineering of pyrrolysyl-tRNA synthetase to genetically encode Ne-(o-azidobenzyloxycarbonyl)lysine for site-specific protein modification”, Chem. Biol. 15, 1187-1197 (2008)
[Incorporation of synthetic amino acids in mammalian cells]
-
Sakamoto, K., Hayashi, A., Sakamoto, A., Kiga, D., Nakayama, H., Soma, A., Kobayashi, T., Kitabatake, M., Takio, T., Saito, K., Shirouzu, M., Hirao, I., and Yokoyama, S., “Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells”, Nucleic Acids Res. 30, 4692-4699 (2002)
[Incorporation of synthetic amino acids in insect cells]
-
Mukai, T., Wakiyama, M., Sakamoto, K., Yokoyama, S., “Genetic encoding of non-natural amino acids inDrosophila melanogaster Schneider 2 cells”, Protein Sci. 19, 440-448 (2010)
[E. coli RFzero strains]
-
Mukai, T., Hayashi, A., Iraha, F., Sato, A., Ohtake, K., Yokoyama, S., Sakamoto, K. “Codon reassignment in the Escherichia coli genetic code”, Nucleic Acids Res. 38, 8188-8195 (2010)
-
Mukai, T., Yanagisawa, T., Ohtake, K., Wakamori, M., Adachi, J., Hino, N., Sato, A., Kobayashi, T., Hayashi, A., Shirouzu, M., Umehara, T., Yokoyama, S., Sakamoto, K., “Genetic-code evolution for protein synthesis with non-natural amino acids”, Biochem. Biophys. Res. Commun. 411, 757-761 (2011)
[Experimental procedures]
-
Hino, N., Sakamoto, K., Yokoyama, S., “Site-specific incorporation of unnatural amino acids into proteins in mammalian cells”, in Unnatural Amino Acids: Methods in Molecular Biology, Volume 794, Part 3, pp. 215-228 (2012)
-
Hino, N., Hayashi, A., Sakamoto, K., Yokoyama, S., “Site-specific incorporation of non-natural amino acids into proteins in the mammalian cells with an expanded genetic code”, Nat. protocol 1, 2957-2962 (2007)