Technologies of CLST
SCENT: Single Cell Expression Technology
Cells in a given population exhibit heterogeneous (mixed) gene expression patterns, suggesting that no single cells are equal. Expression profiling of individual cells is therefore essential for the understanding of gene regulation and can detect aberrant cell types in a seemingly healthy population.
In DGT, we established a Single Cell Expression Technology, or SCENT, that streamlines multiple laborious steps into one simple workflow – further incorporating single cell experiments and expression data with a centralized database for easy deposition and dissemination of single cell expression data

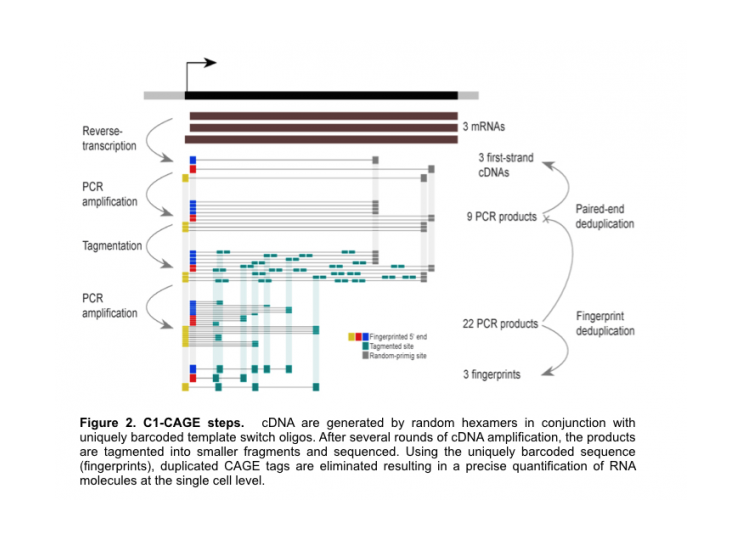
SCENT relies on the microfluidics Fluidigm C1™ system to capture single cells, followed by General Electrics’ (GE) InCell6000 imaging system for quantification of cellular features (e.g. GFP expression levels). Once the cells are captured, SCENT executes either SMARTer® RNA-seq protocol or C1-CAGE, a RNA expression profiling method developed in DGT (link). In comparisons to SMARTer® RNA-seq, C1-CAGE employs random hexamers to yield strand-specific 5’-end cDNA tags to quantify promoter activities near the Transcription Start Sites (TSS). The Unique Molecular Identifier (UMI) encoded in the template switching oligo allows for the precise quantification of CAGE tags - unbiased to the inefficiencies introduced during the amplification steps (Figure 2).
SCENT is a versatile and scalable platform and it is currently being applied in various biological systems inside RIKEN:
- Dynamic drug response
- Cell reprogramming
- Cancer/cell-cycle
- Neurobiology
- Aging
The script to run C1 CAGE can be found here.