Technologies & Facility
Technologies
Division of Structural and Synthetic Biology
Incorporation of synthetic amino acids into proteins at specific sites
This technology is about site-specific incorporation of synthetic amino acids into proteins in animal cells and E. coli cells.
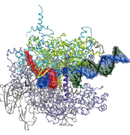
X-ray crystallography for macromolecular complexes
Organismal bodies and biogenic activities are sustained by various macromolecules including proteins, DNA, and RNA. These bio-molecules express their functions or activities by interacting with each other or by forming gigantic molecular complexes. Studying the three-dimensional structures of these bio-molecules allows us to understand their action mechanism. We employ X-ray crystallography for the structural analyses. The obtained three-dimensional structures provide not only insights into detailed molecular mechanisms, but also firm bases for applied researches such as drug development.
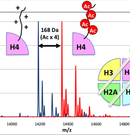
Epi-nucleosome’ technologies (Precise reconstitution of epigenetic nucleosomes)
Epigenetic information is ‘added’ onto genetic information, which regulates gene expression of individual cell. This ‘additional’ information is substantialized by chemical modification onto nucleosome that is composed of DNA and histone proteins. The modification of the histone proteins is recorded on the nucleosome, as combinations of the kind of modifications (acetylation, methylation, phosphorylation etc.), and the kind of the subunit and residues modified. These combinatorial information is maintained, recognized and converted in the cell. Although the regulatory mechanisms of epigenetics have been extensively analyzed at the cellular level, biochemical analyses mostly remained at those using modified peptides as pseudo-substrate of chromatin.
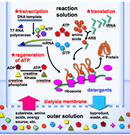
Cell-Free Protein Production System
Cell-free protein production system consists of RNA polymerase, transfer RNAs (tRNAs), aminoacyl-tRNA synthetases, translation factors, ribosomes, and DNA for the template of a target protein.
Protein Functional and Structural Biology Team,Nonnatural Amino Acid Technology Team
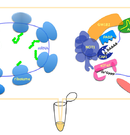
Cell-free system that recapitulates miRNA-mediated mRNA regulation.
Regulation of translation and mRNA degradation is based on a complex interplay of canonical factors, diverse RNA-binding proteins, and non-coding regulatory RNAs (e.g. microRNAs). We have established an in vitro experimental system to recapitulate mechanisms of miRNA-mediated post-transcriptional control by utilizing a cell-free translation technique.
Production and Crystallization of Membrane Proteins
Membrane proteins play indispensable roles in the living cells, such as signal transduction between outside and inside of the cell (receptors), solute transporters, ion channels, and so on, thus many of them have been drug discovery targets.
Structural analysis of bio-macromolecular complexes in electron cryo-microscopy
Structural analysis of bio-macromolecular complexes in electron cryo-microscopy (cryo-EM) is a relatively new method in structural biology.There are two major methods in the cryo-EM, one is called cryo-electron tomography and the other is single particle cryo-EM.
Division of Bio-Function Dynamics Imaging
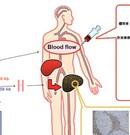
PET imaging
Two dedicated PET scanners (Fig.1), two combined PET/CT scanners (Fig. 2) and one CT scanner (Fig. 3) are installed in RIKEN CLST-DBDI Kobe.
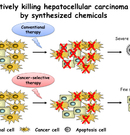
Technique to kill selectively hepatocellular carcinoma cells by synthesized chemicals
Hepatocellular carcinoma (HCC) represents approximately 85% of all primary liver cancer and is one of the most common malignancies worldwide, especially in Eastern Asia.
Technique to detect tissue fibrogenesis using the antibodies against TGF-β LAP degradation products
The most fibrogenic cytokine, TGF-β, is produced as a latent complex, in which active TGF-β is wrapped by latency associated protein (LAP), preventing TGF-β from binding its receptor on target cells.
Analysis of protein-crosslinking activity in the nucleus
Transglutaminases covalently crosslink proteins between Lys and Gln residues.
The “KOKORO scale”, a system to assess human mood or mental state.
The “KOKORO scale”, which was developed by Cellular Function Imaging Team is a system in which individuals can input mood information simply with a touch panel or other device. It utilizes four-quadrant matrix*1 with measures of mood including feelings of security and anxiety, feelings of excitement and irritability, and level of motivation (feeling considerable motivation and feeling no motivation) on horizontal and vertical axes.
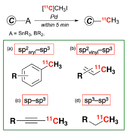
Rapid C-[11C]methylation
To acquire more accurate PET image, it is important that the PET probe should be stable, both chemically and metabolically, in the imaging time window.
Division of Genomic Technologies
CAGE : A method for genome-wide identification of transcription start sites
Cap Analysis Gene Expression is a technology focused on the 5' sequence analysis, capturing capped 5' end of RNAs, originally developed by RIKEN in 2003. TSS sequence is quite informative, since you can find corresponding promoter sequence and thereby, you can also predict transcription factor regulation for each transcript.
Transcriptome Technology Team,Genomics Miniaturization Technology Unit
ZENBU: Interactive visualization and analysis genome browser system
The ZENBU Genome Browser was developed at RIKEN for the FANTOM 5 project. ANTOM5 needed a more powerful genome browser system which could manage CAGE & RNAseq transcriptomics data from 1000s of different cell and tissue samples. ZENBU is a next-generation Sequencing (NGS) focused genome browser for transcriptomics and epigentics data.
Technologies for FANTOM Web Resources
Recent genomics data tends to be large-scale and diverse, for example, tables, texts, DNA/RNA sequences, images, and so forth. As such, well-organized visualization and databases systems are required to access the data. In the FANTOM5 project, we created a series of database systems for this purpose.
ACCIS: Automated Cell Culturing System
Cells possess all of its genetic materials required to function (e.g. DNA and RNA) and these genetic materials can be extracted and profiled to understand the cellular function. Further, because in vitro cells can be maintained with precise step-by-step procedures, we developed an Automated Cell Culturing System (ACCIS) in order to screen for cellular functions by genetic perturbation (e.g. Loss-of-function or gain-of-function).
SCENT: Single Cell Expression Technology
DGT established a Single Cell Expression Technology, or SCENT, that streamlines multiple laborious steps into one simple workflow – further incorporating single cell experiments and expression data with a centralized database for easy deposition and dissemination of single cell expression data
Genomics Miniaturization Technology Unit,Large Scale Data Managing Unit,Cell Conversion Technology Team
SINEUPs: A new class of translation regulating RNA
SINEUP is a class of novel non-coding RNAs (ncRNAs), which was reported in 2012. Since they can be used as a tool to stimulate protein translation of mRNA of interest, broad applications of it is expected from research reagent to therapeutic.
SmartAmp: Smart Amplification Process
The Smart Amplification process (SmartAmp) method uses isothermal nucleic acid amplification technology and employs a single reaction temperature (60-67ºC), which translates into simple and less expensive instrumentation