Unit Leader
Exploring the RNP complexes involved in the regulation of protein synthesis.
* Due to the reorganization starting as new centers in April 2018, the research activities of this laboratory are handed over to the Center for Biosystems Dynamics Research. As for the latest information, please see the following URL below.
> The webpage of Laboratory for Nonnatural Amino Acid Technology, Center for Biosystems Dynamics Research

Unit Leader
Motoaki Wakiyama
1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan
![]()
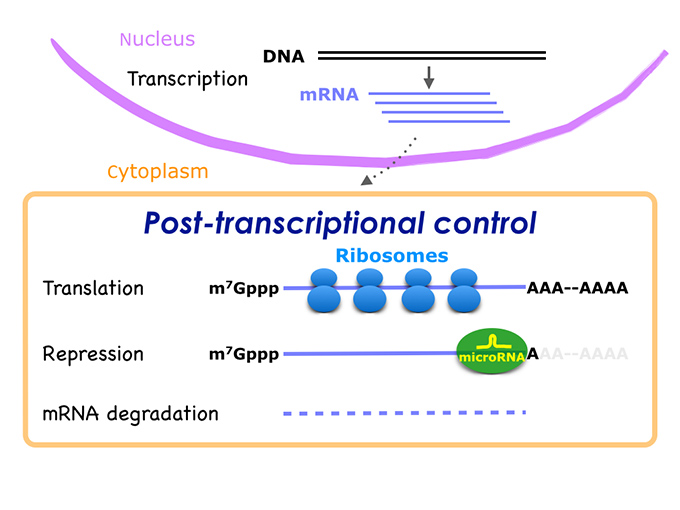
Control of cellular function and cell fate determination is dependent on the regulation of gene expression. The eukaryotic gene expression was thought to be mainly regulated at transcription level. However, based on the accumulation of the knowledge in microRNA function, it is now believed that post-transcriptional control of gene expression is much more diverse and intricate than previously thought, and is extensively involved in numerous diseases. Post-transcriptional gene regulation is mediated by a number of RNA-binding proteins (RBP), RBP-binding proteins and non-coding regulatory RNAs. These factors associate with the target mRNA molecule and form a huge mRNP complex. We aim to uncover the molecular mechanism of mRNA fate determination, by developing a novel method for the investigation of the architecture and function of mRNPs.


CLST was reorganized into three centers according to the RIKEN 4th Medium-Term Plan from April 1, 2018.
> The webpage of Laboratory for Nonnatural Amino Acid Technology, Center for Biosystems Dynamics Research [http://www.bdr.riken.jp/en/research/labs/sakamoto-k/index.html]
> If you continue to browse this site, click here.