Unit Leader

We try to develop molecules that intervene in cellular pathways and thus modulate cell functions.
* Due to the reorganization starting as new centers in April 2018, this laboratory is now belong to the Center for Biosystems Dynamics Research. As for the latest information, please see the following URL below.
> The webpage of Laboratory for Advanced Biomolecular Engineering, Center for Biosystems Dynamics Research
Unit Leader
Shunsuke Tagami
Room C516, Central Building, Yokohama Campus
1-7-22 Suehiro-cho,Tsurumi-ku, Yokohama 230-0045
TEL: 045-503-9205
![]()
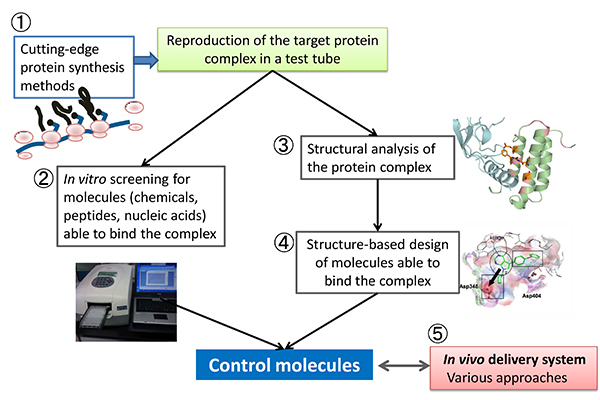
This unit aims to engineer functional bio-macromolecules applicable to medical and basic biological researches, by integrating technologies in synthetic and structural biology including 1) in vitro reconstruction of target protein complexes, 2) in vitro evolution of peptide/RNA, 3) protein X-ray crystallography, and 4) structure based protein/drug engineering. Delivery systems for the engineered molecules will be also developed in collaboration with the Molecular Network Contraol Imaging Unit.
CLST was reorganized into three centers according to the RIKEN 4th Medium-Term Plan from April 1, 2018. For the latest information of Molecular Network Control Factors Development Unit, please visit the following websites.
> The webpage of Laboratory for Advanced Biomolecular Engineering, Center for Biosystems Dynamics Research [http://www.bdr.riken.jp/en/research/labs/tagami-s/index.html]
> If you continue to browse this site, click here.